Introduction
Prostate cancer (PCa) is the eighth leading cause of death among older men worldwide. According to the 2022 GLOBOCAN report, this malignancy accounted for 397,430 deaths globally, with 61,957 occurring in Latin America and the Caribbean1,2. In addition, PCa is highly metastatic, has a low cure rate, and exhibits significant resistance to treatment. These challenges have prompted the development of more sensitive and specific laboratory techniques to enable early detection. In this context, biomarkers have emerged as promising tools to improve diagnostic accuracy2–4.
At present, prostate-specific antigen (PSA) is the most widely used biomarker for early PCa detection. While PSA testing has increased detection rates, it is also associated with overdiagnosis, leading to unnecessary biopsies. These invasive procedures carry risks such as infection, hematuria, hematochezia, and urinary retention5–7.
Given the previously mentioned risks, it is crucial to explore other biomarkers to improve diagnostic specificity and reduce unnecessary biopsies. In recent years, small non-coding RNAs (ncRNAs) have been identified as key players in tumorigenesis, progression, and prognosis in various cancers. PIWI-interacting RNAs (piRNAs), a class of ncRNAs, 24-31 nucleotides in length, interact with PIWI proteins and play a fundamental role in germline maintenance. piRNAs exhibit specific characteristics, including a uridine signature at their 5’ end, adenosine at the 10th position, and a stable 2’-O-methylation modification at the 3’ end. They are associated with PIWI proteins, which are involved in transposon silencing, histone modification, heterochromatin formation, and genomic integrity regulation through transcriptional and post-transcriptional mechanisms8–2.
Despite extensive research on the potential use of piRNAs as biomarkers and therapeutic targets in PCa, their clinical utility remains unclear3. Therefore, this systematic review aims to evaluate the role of piRNAs in PCa, focusing on their potential as biomarkers and therapeutic targets. The goal is to provide clarity on their applicability in clinical practice.
Methods
The reporting of this review followed the guidelines set by the 2020 preferred reporting items for systematic reviews and meta-analyses Statement (Fig. 1)4,5.
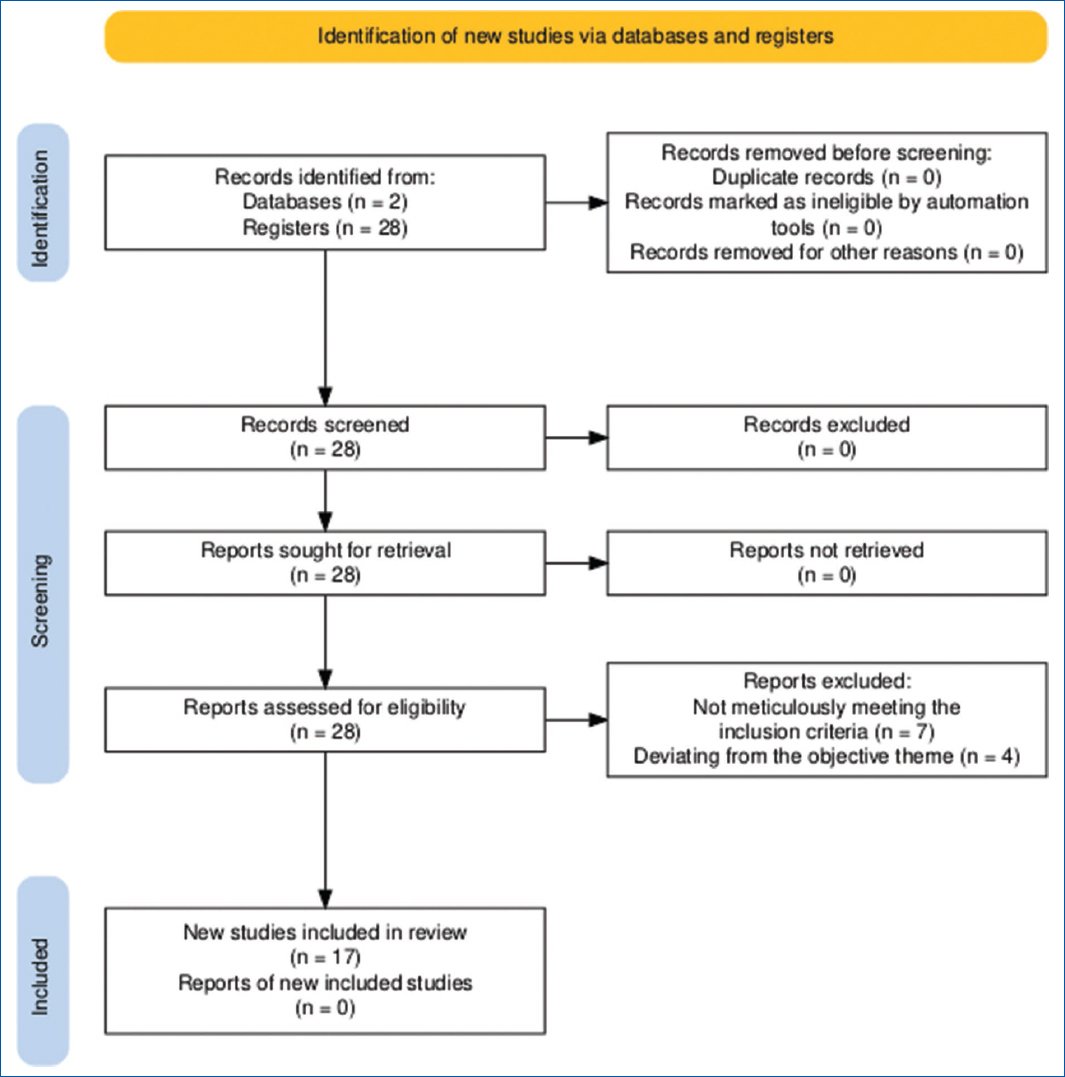
Figure 1. Preferred reporting items for systematic reviews and meta-analyses flow chart4,5.
Literature search strategy
To systematically evaluate the role of piRNAs and PIWI-like proteins in the diagnosis, prognosis, and treatment of PCa, a comprehensive literature search was conducted using the PubMed database. The search strategy aimed to identify relevant studies focused on the clinical implications of piRNAs and PIWI-like proteins in PCa.
Search terms, inclusion, and exclusion criteria
The search terms used were: ([piRNA] OR [PIWI-like protein]) AND (PCa).
Studies were included based on the following inclusion criteria
- Study type: Original research articles, clinical trials, and systematic reviews
- Population: Studies involving human subjects with PCa
- Intervention/focus: Research exploring the role of piRNAs, PIWI-like proteins, or their related pathways as biomarkers or therapeutic targets in PCa
- Language: Articles published in English
- Publication date: No restrictions were applied based on the date of publication
- Outcome: Studies reporting outcomes related to the role of piRNAs or PIWI-like proteins in the diagnosis, prognosis, or treatment of PCa.
Exclusion criteria were as follows
- Study type: Case reports, case series, cohort studies, case–control studies, editorials, letters to the editor, and non-peer-reviewed articles
- Population: Studies involving animal models or non-human subjects
- Focus: Research not specifically addressing piRNAs or PIWI-like proteins in PCa
- Language: Articles not available in English
- Outcome: Studies that did not provide specific outcomes related to piRNAs or PIWI-like proteins in PCa.
Data extraction and management
THE DATA EXTRACTED FROM THE SELECTED STUDIES INCLUDED THE FOLLOWING
- Study characteristics: Principal author, year of publication, study design, sample size, and characteristics of the patients
- Intervention details: Focus on piRNAs or PIWI-like proteins, including molecular pathways and experimental techniques used
- Outcomes: Primary outcomes related to the role of piRNAs and PIWI-like proteins in PCa, such as diagnostic value, prognostic markers, therapeutic potential, and their effects on tumor progression.
Two independent reviewers conducted the data extraction to ensure accuracy and consistency, resolving any discrepancies through discussion or consultation with a third reviewer.
Quality assessment
The quality of the included studies was assessed using appropriate methodological tools based on the study design, specifically the Newcastle-Ottawa scale (Table 1)6. This evaluation focused on the risk of bias, study validity, and the overall reliability of the findings.
Table 1 Newcastle-Ottawa scale6
| Principal Author | Year | Selection | Comparability | Outcome | Total Score |
|---|---|---|---|---|---|
| Ben S | 2024 | 4 | 2 | 3 | 9 |
| Peng Q | 2024 | 4 | 2 | 3 | 9 |
| Ben S | 2023 | 3 | 1 | 2 | 6 |
| Kocic G | 2022 | 3 | 1 | 2 | 6 |
| Peng Q | 2021 | 4 | 2 | 3 | 9 |
| Markert L | 2021 | 3 | 2 | 3 | 8 |
| Zhang L | 2020 | 4 | 2 | 3 | 9 |
| Qi T | 2020 | 3 | 2 | 3 | 8 |
| Zhang L | 2020 | 4 | 2 | 3 | 9 |
| Tosun H | 2019 | 4 | 2 | 3 | 9 |
| Zuo Y | 2019 | 3 | 2 | 3 | 8 |
| Baumann B | 2019 | 4 | 1 | 3 | 8 |
| Yang Y | 2015 | 4 | 2 | 2 | 8 |
| Ben S | 2024 | 4 | 2 | 3 | 9 |
| Peng Q | 2024 | 4 | 2 | 3 | 9 |
| Zhang L | 2020 | 4 | 2 | 3 | 9 |
| Zuo Y | 2019 | 3 | 1 | 3 | 7 |
Data synthesis
A narrative synthesis was conducted to integrate and summarize the findings from the reviewed studies. The synthesis focused on discussing their potential as diagnostic biomarkers and therapeutic targets, as well as their role in predicting clinical outcomes in PCa.
Results
In this systematic review, we synthesized findings from 17 studies that investigated the roles of piRNAs and PIWI-like proteins in PCa (Table 2). The results reveal a multifaceted involvement of piRNAs in various stages of PCa progression, metastasis, therapeutic resistance, and potential clinical applications as diagnostic and prognostic biomarkers.
Table 2 Characteristics of the reviewed studies
| Principal Author | Year | Main Result | Type of Study | # of Patients |
|---|---|---|---|---|
| Ben S | 2024 | piRNA PROPER promotes oncogenesis by degrading DUSP1 via m6A-mediated circularization | Experimental (in vitro, in vivo) | 85707 |
| Peng Q | 2024 | piR-4447944 promotes castration-resistant growth and metastasis by inhibiting NEFH | Experimental (in vitro, in vivo) | 19 |
| Ben S | 2023 | BPA promotes prostate cancer invasion via piR-sno48-mediated suppression of GSTP1 | Experimental (in vitro) | PC-3 cells |
| Kocic G | 2022 | NA-TLRs and nucleases play a role in PCa immune escape and xenophagy | Observational | N/A |
| Peng Q | 2021 | Urinary EVs piRNAs can serve as non-invasive biomarkers for PCa | Case-control | 35 |
| Markert L | 2021 | Small RNAs differentiate benign from malignant prostate diseases using machine learning | Case-control | 53 |
| Zhang L | 2020 | piR-001773 and piR-017184 promote PCa by regulating PCDH9 and AKT | Experimental (in vitro, in vivo) | N/A |
| Qi T | 2020 | piR-19166 inhibits metastasis by targeting CTTN and MMP pathways | Experimental (in vitro, in vivo) | N/A |
| Zhang L | 2020 | piR-31470 silences GSTP1 via DNA methylation, promoting PCa progression | Experimental (in vitro) | N/A |
| Tosun H | 2019 | PIWIL2 serum levels are a prognostic indicator for advanced PCa | Prospective | 60 |
| Zuo Y | 2019 | Three piRNAs predict PCa recurrence and are associated with Gleason score | Transcriptome analysis | N/A |
| Baumann B | 2019 | Vitamin D regulates piRNA expression in PCa epithelium, suggesting a protective role | Observational | N/A |
| Yang Y | 2015 | PIWIL2 modulates PCa invasion via MMP-9 and EMT pathways | Experimental (in vitro) | 30 |
| Ben S | 2024 | piRNA PROPER promotes oncogenesis by degrading DUSP1 via m6A-mediated circularization | Experimental (in vitro, in vivo) | 85707 |
| Peng Q | 2024 | piR-4447944 promotes castration-resistant growth and metastasis by inhibiting NEFH | Experimental (in vitro, in vivo) | 19 |
| Zhang L | 2020 | piR-31470 silences GSTP1 via DNA methylation, promoting PCa progression | Experimental (in vitro) | N/A |
| Zuo Y | 2019 | Three piRNAs predict PCa recurrence and are associated with Gleason score | Transcriptome analysis | N/A |
|
PCa: prostate cancer; CRPC: ccastration-resistant prostate cancer; BPA: bisphenol A.; NA-TLRs; nucleic acid-Sensing toll-like receptors; EV: extracellular vesicles; PC-3: prostate cancer cell line 3; GSTP1: glutathione S-transferase Pi 1; MMP: matrix metalloproteinase; CTTN: cortactin; EMT: epithelial-mesenchymal transition. |
||||
Key piRNAs were identified as oncogenic drivers of PCa. For instance, Ben et al. (2024) demonstrated that piRNA PROPER promotes prostate tumorigenesis by targeting DUSP1 degradation through m6A-mediated RNA circularization, which in turn activates the p38 mitogen-activated protein kinase (MAPK) pathway, contributing to tumor progression7. Similarly, Peng et al. (2024) highlighted piR-4447944 as a critical regulator of castration-resistant PCa (CRPC), where it suppresses neurofilament heavy polypeptide (NEFH), a tumor suppressor gene, through its interaction with PIWIL2, facilitating metastasis8. These findings are consistent with Zhang et al. (2020), who showed that piR-001773 and piR-017184 promote PCa by downregulating PCDH9, leading to the activation of the PI3K/AKT signaling pathway1.
In terms of metastasis, several studies revealed that piRNAs play crucial roles in enhancing the invasive potential of PCa cells. In a study conducted by Ben et al., 2024, piR-sno48 was shown to enhance the migratory properties of PCa cells under the influence of bisphenol A (BPA)7. Conversely, Qi et al. (2020) found that piR-19166 suppresses cell migration and metastasis by targeting the CTTN/MMP pathway, suggesting that piRNAs may have both pro-metastatic and anti-metastatic roles depending on the molecular context10.
Furthermore, the epigenetic modulation of key tumor suppressor genes by piRNAs is evident. Zhang et al. (2020) demonstrated that piR-31470 epigenetically silences GSTP1 through DNA methylation, which increases susceptibility to oxidative stress and promotes tumor progression1. This aligns with Baumann et al. (2019), In their study, Baumann et al. (2019) explored the expression of piRNAs in normal prostate epithelium and the potential regulatory effects of Vitamin D on these small ncRNAs. The findings revealed an unexpected abundance of piRNAs in normal prostate epithelium, along with the presence of PIWI-like proteins, which are essential for piRNA function. Notably, higher prostatic levels of Vitamin D were associated with increased expression of piRNAs. Vitamin D receptor (VDR) chromatin immunoprecipitation sequencing identified VDR binding sites near several genes involved in ncRNA biogenesis and those regulating translation and differentiation. These results suggest that Vitamin D may upregulate piRNA expression in the prostate epithelium, potentially influencing gene regulatory mechanisms involved in PCa progression. The study highlights the possibility that external factors, such as Vitamin D levels, can modulate piRNA-mediated epigenetic changes. This aligns with the concept that environmental factors can impact the expression and function of piRNAs, thereby affecting cancer biology9.
piRNAs also show potential as biomarkers for PCa. Peng et al. (2021) identified urinary piRNAs, including novel_pir158533 and hsa_piR_002468, as non-invasive biomarkers for PCa diagnosis with high accuracy3. Other studies, such as Tosun et al. (2019), explored PIWIL2 expression as a serum marker, although its diagnostic utility was limited, its prognostic value in advanced disease was evident20.
Discussion
The emerging evidence underscores the critical role that piRNAs and PIWI-like proteins play in the biology of PCa. These small ncRNAs operate through perse molecular mechanisms, influencing key processes such as tumorigenesis, metastasis, epigenetic regulation, and resistance to therapy. A consistent theme across multiple studies is the dual role of piRNAs as either oncogenic drivers or tumor suppressors, depending on their molecular targets and the cellular context. This duality highlights the complexity of piRNA biology in PCa.
One of the most prominent findings across the literature is the involvement of piRNAs in driving tumorigenesis and promoting metastatic behavior. The studies conducted by Ben et al. (2024) and Peng et al. (2024) exemplify how piRNAs such as PROPER and piR-4447944 contribute to PCa progression by regulating key tumor suppressor genes, such as DUSP1 and NEFH. These piRNAs mediate complex interactions at the post-transcriptional level, often affecting multiple signaling pathways, including p38 MAPK and PI3K/AKT. This interplay between piRNAs and signaling cascades evidence their potential as therapeutic targets for intervention7,8.
In addition to promoting tumorigenesis, piRNAs also play an integral role in metastatic processes. The upregulation of piRNAs, such as piR-sno48 in response to environmental carcinogens, such as BPA suggests a direct link between external factors and piRNA-mediated cancer progression2. Moreover, the suppression of metastatic potential by piRNAs such as piR-19166 points to the therapeutic promise of modulating piRNA activity. This contrasting role of piRNAs in either enhancing or suppressing metastasis adds to the complexity of their regulatory functions in PCa and highlights the importance of context when considering piRNAs as therapeutic targets10.
The epigenetic regulation of gene expression is another key mechanism through which piRNAs influence PCa progression. As demonstrated by Zhang et al. (2020), the ability of piRNAs, such as piR-31470 to silence tumor suppressor genes, such as GSTP1 through DNA methylation illustrates their critical role in modulating the cancer epigenome. These findings align with the growing body of evidence that suggests piRNAs could be harnessed as therapeutic agents, either to restore tumor suppressor gene function or to inhibit oncogenic piRNA activity6,20,21.
The potential of piRNAs as biomarkers for PCa diagnosis and prognosis is particularly promising. In the study by Peng et al. (2021), the researchers explored the potential of piRNAs in urinary extracellular vesicles (EVs) as non-invasive biomarkers for the diagnosis of PCa. Four specific piRNAs – novel_pir349843, novel_pir382289, novel_pir158533, and hsa_piR_002468 – were found to be significantly overexpressed in the PCa group compared to the healthy control group. The findings were validated in a larger cohort consisting of 30 PCa patients and 10 healthy controls using reverse-transcription polymerase chain reaction. The diagnostic value of these piRNAs was assessed through the area under the curve (AUC), which showed promising results: 0.723 for novel_pir158533, 0.757 for novel_pir349843, 0.777 for novel_pir382289, 0.783 for hsa_piR_002468, and a combined AUC of 0.853 when all four piRNAs were considered together. These values suggest a strong potential for these piRNAs to serve as diagnostic biomarkers. This study provides compelling evidence that specific piRNAs present in urinary EVs could serve as non-invasive biomarkers for the early diagnosis of PCa, offering a promising alternative to invasive diagnostic procedures21. In addition, the prognostic value of piRNAs in predicting disease progression, as shown by studies, such as Zuo et al. (2019), further supports their clinical utility in managing PCa21,22.
Conclusion
piRNAs and PIWI-like proteins are emerging as crucial components in the regulation of PCa, influencing oncogenesis, metastasis, and resistance to therapy. These molecules can act as either oncogenes or tumor suppressors depending on the cellular context, making them promising targets for novel therapeutic strategies. piRNAs also show potential as non-invasive biomarkers, as urinary piRNAs may provide a valuable alternative to present invasive diagnostic methods, offering a pathway to more accurate and earlier diagnosis of PCa. The therapeutic modulation of piRNAs presents a significant opportunity for advancing PCa treatment, particularly in cases of CRPC where treatment options are limited. Further research into their molecular mechanisms and environmental interactions is essential to fully comprehend their potential as both therapeutic targets and biomarkers in the clinical management of PCa.
Funding
The authors declare that they have not received funding.
Conflicts of interest
The authors declare no conflicts of interest.
Ethical considerations
Protection of humans and animals. The authors declare that no experiments involving humans or animals were conducted for this research.
Confidentiality, informed consent, and ethical approval. The study does not involve patient personal data nor requires ethical approval. The SAGER guidelines do not apply.
Declaration on the use of artificial intelligence. The authors declare that no generative artificial intelligence was used in the writing of this manuscript.
